This guest post is by Jerome Kelleher, on the preprint by Kelleher, Etheridge, and McVean titled “Efficient coalescent simulation and genealogical analysis for large sample sizes” available here from biorXiv.
In this post we summarise the main results of our recent bioRxiv preprint. We’ve left out a lot of important details here, but hopefully this summary will be enough to convince you that it’s worth reading the paper!
Coalescent simulation is a fundamental tool in modern population genetics, and a large number of packages exist to simulate various aspects of the model. The basic algorithm to simulate the coalescent with recombination was defined by Hudson in 1983, who also published the classical ms simulation program in 2002. Programs such as ms based on Hudson’s algorithm perform poorly for longer sequences, making simulation of chromosome sized regions under the influence of recombination impossible. The Sequentially Markov Coalescent (SMC) approximates the coalescent with recombination by assuming that each marginal genealogy depends only on its predecessor, making simulation much more efficient. The SMC can be a poor approximation when long range linkage information is important, however, and current simulations do not scale well in terms of sample size. Population-scale sequencing projects currently under way mean there is an urgent need for accurate simulations of hundreds of thousands of genomes.
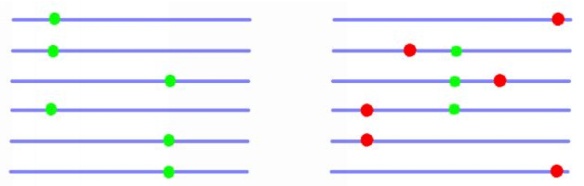
We present a new formulation of Hudson’s simulation algorithm that solves these issues, making chromosome-scale simulation of the exact coalescent with recombination for hundreds of thousands of samples possible for the first time. Our approach begins by defining the genealogies that we are constructing in terms of integer vectors of a specific form, which we refer to as `sparse trees’. We generate recombination and common ancestor events in the same manner as the classical methods, but our approach to constructing marginal genealogies is quite different. When a coalescence within a marginal tree occurs, we store a tuple consisting of the left and right coordinates of the overlapping region, the parent and child nodes (which are integers), and the time at which the event occurred. We refer to these tuples as `coalescence records’, and they provide sufficient information to recover all genealogies after the simulation has completed. We implemented these ideas in a simulator called msprime, which we compared with the state of the art. For a fixed sample size of 1000 and increasing sequence length (with human-like recombination parameters), we found that msprime is much faster than comparable exact simulators and, surprisingly, is competitive with approximate SMC simulators. Even more surprisingly, we found that for a fixed sequence length of 50 megabases and increasing sample size, msprime was much faster than any existing simulator for large samples.
Storing the output of the simulations as coalescence records has major advantages. Because parent-child relationships shared by adjacent trees are stored only once, the correlation structure of the sequence of genealogies is explicit and the storage requirements minimised. To illustrate this, we ran a simulation of a 100 megabase chromosome with a roughly human recombination rate for a sample of 100,000 individuals. This simulation ran in about 6 minutes on a single CPU thread and used around 850MB of RAM. The resulting coalescence records required 88MB using msprime’s native HDF5 based storage format. Storing the same genealogies in Newick format requires around 3.5TB.
Highly compressed representations of data usually come at the cost of increased access time. In contrast, we can retrieve complete genealogical information from coalescence records many times faster than is possible using existing Newick-based methods. We provide a detailed listing of an algorithm to sequentially recover the marginal genealogies from a set of coalescence records and show that this algorithm requires constant time to transition between adjacent trees. This algorithm has been implemented as part of msprime’s Python API, and required around 3 seconds to iterate over all 1.1 million trees generated by the simulation above. We compared this performance to several popular tree processing libraries, and found that the fastest would require an estimated 38 days to parse the same set of trees in Newick format. Thus, in this example, by using msprime’s storage format and API we can store the same set of trees using around forty thousand times less space and parse them around a million times more quickly than Newick based methods.
We can also store mutational information in a natural and efficient way. If we have an infinite sites mutation that occurs on the parent branch of a particular node at a particular position on the sequence, then we simply store this (node, position) pair. This leads to a very compact representation of the combined genealogical and mutational state of a sample. We simulated 1.2 million infinite sites mutations on top of the genealogies generated earlier, which resulted in a 102MB HDF5 file containing the coalescence records and mutations. In contrast, the corresponding text haplotypes consumed 113GB of storage space. Associating mutations directly with tree nodes also allows us to perform some important calculations efficiently. We describe an efficient algorithm to count the total number of leaf nodes from a particular set below each node in the tree as we iterate over the sequence. This algorithm allows us to (for example) calculate allele frequencies within specific subsets in constant time. Many other applications of these techniques are possible.
The availability of faster and more accurate simulations may lead to interesting new applications, and so we conclude by discussing some potential applications of our work. Of particular interest is the possibility of inferring a set of coalescence records from real biological data, obtaining a compressed representation that can be efficiently processed. This is a very interesting and promising direction for future research.